We study how populations adapt to changes in their environment. We are particularly interested in populations of bacterial and viral pathogens, and how their evolution impacts infectious disease dynamics. We use mathematical modelling, experiments, and statistical inference in our research.
Research interests and current projects.
Bacterial evolution of antibiotic resistance presents a formidable challenge for public health. As discovery of new antibiotic classes has slowed down, an important question is how best to use existing antibiotics. Can we dose these drugs in a way that is clinically effective, but avoids evolution of resistance?
We are currently tackling this question at its most basic level – examining the response of bacterial populations to antibiotics in vitro and in mathematical models. We are especially interested in how resistant mutants first arise and spread in an initially antibiotic-sensitive population. Importantly, rare resistant lineages are subject to strong demographic stochasticity, and are at risk of loss even if they are selectively favoured.
We experimentally demonstrated the importance of stochastic loss of rare resistant cells in recent work with Prof. Craig MacLean at the University of Oxford. Our results suggested that antibiotic doses that are too low to clear an established resistant population may nonetheless prevent rare resistant lineages from getting off the ground in the first place.

We are continuing this experimental work at the University of Edinburgh, with a particular interest in how ecological effects – such as competition and protection by surrounding cells – impact the emergence of antibiotic resistance. In parallel, we are developing and analysing mathematical models of bacterial population dynamics in their changing environment. We will use these models to generate predictions – for instance, to explore optimal antibiotic dosing – and to help determine which parameters are critical to measure experimentally.
- Alexander & MacLean, PNAS, 2020
Group members currently working on this topic: Grace Taylor-Joyce (experiments), Pierre Lafont (mathematical models)
Mutations (heritable changes to an organism’s genome) are the ultimate source of genetic variation fuelling evolution. While most mutations are deleterious, rare beneficial mutations allow organisms to adapt to environmental challenges. Thus, understanding how – and how often – mutations arise is critical to predicting how populations evolve.
Typically, population genetic and evolutionary models represent mutations as instantaneous events converting an individual from one “type” to another. However, reality is more complicated: heritable changes to the DNA are the net result of multiple intracellular processes. Moreover, a change of genotype may have a delayed effect on the phenotype. And finally, these processes may vary among individuals and over time, particularly under exposure to environmental stressors. We are particularly interested in how these processes play out in bacteria, which are both experimentally tractable and relevant for medical concerns (e.g. antibiotic resistance evolution).
Understanding the impacts of these mutational complexities is an ongoing topic of research in our group. We are primarily using modelling and simulation techniques to ask:
- If we incorporate more realistic descriptions of the mutation process into our models, how does this affect the predicted population dynamics and evolution?
- How do these deviations from standard model assumptions impact estimates of mutation rates from standard experimental assays?
Our models are also closely informed by collaborations with other experimental labs investigating bacterial responses on the single-cell level.
- Alexander, Mayer & Bonhoeffer, Molec. Biol. Evol., 2017
- Sun, Alexander et al., PLoS Biol., 2018
- Lansch-Justen et al., bioRxiv preprint, 2023
Group members currently working on this topic: Lucy Lansch-Justen
When susceptible hosts are exposed to pathogens, their chances of getting infected typically increase with the dose, i.e. number of pathogens or inoculum size, to which they are exposed. Dose-response models are used to describe the relationship between dose and probability of host infection (in theory) or proportion of hosts in which an infection is observed to establish (in experimental or natural infections). The simplest, widely used theoretical model is based on the "independent action hypothesis", which posits that each individual pathogen (e.g. bacterial cell or virion) has a certain probability of establishing infection, independently of how many other pathogens are present in the dose. This leads to a particularly simple mathematical formulation. However, competitive or cooperative interactions within a pathogen population in the process of infecting a host would lead to deviations from the independent action model.
In current work, we are investigating:
- How well the independent action model fits experimental infection data, and whether particular factors are associated with goodness-of-fit across host/pathogen systems
- How different dose-response models describing host infection impact spread of infectious disease (epidemiology) within a host population
Group members currently working on this topic: Kiran Wadhawan
While our work focuses on microbial pathogens, the question of whether populations can survive environmental challenges arises in much broader contexts, from conservation to agriculture to public health. These diverse applications share a common interest in understanding how populations may adapt to new environments, and ultimately, how we might manipulate their environment to achieve a desired outcome.
“Evolutionary rescue” refers to rapid adaptation that prevents extinction of a population facing a severe environmental change. Evolution of drug resistance in pathogens or cancerous cells, where drug treatment represents the environmental change, is one example of this phenomenon. Evolutionary rescue is also of interest in conservation, despite having the opposite goal of protecting target populations. With Dr. Guillaume Martin (Université Montpellier) and others, we reviewed theoretical work on evolutionary rescue across disciplines. This uncovered different mathematical modelling approaches and a largely disconnected body of literature, despite examining similar population dynamics and asking similar questions. A unified perspective on evolutionary rescue – aiming to draw insights from across different biological systems and understand what makes them similar or different – continues to motivate our current work.
“Evolutionary emergence” refers to the closely related phenomenon where a few individuals invade a new habitat to which they are initially poorly adapted. In order to expand and establish a stable population, they must gain adaptive mutations. One example from epidemiology is the situation where pathogens “invade” a new host species, as in a zoonotic spillover event. We modelled this process in past work with Prof. Troy Day at Queen’s University.

- Alexander & Day, J. R. Soc. Interface, 2010
- Alexander, Adv. Appl. Prob., 2013
- Alexander, Martin et al., Evol. Appl., 2014
We are broadly interested in infectious disease dynamics and the ecology and evolution of pathogens, and welcome project proposals within these general topics.
Particular areas of interest and past projects include:
Viral adaptation to host environmental challenges
Due to their large population sizes, short generation times, and high mutation rates, viruses have a tremendous capacity to evolve. This scope for evolution has profound consequences in chronic viral infections, for instance by HIV, Hepatitis B, and Hepatitis C in humans. These viruses can rapidly adapt to challenging within-host environments, such as immune responses and drug treatments. This can prevent clearance of the infection and impact the infected person’s health.
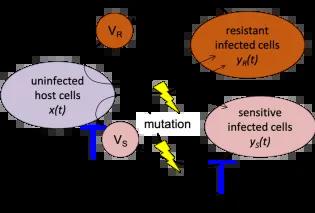
In past work with Prof. Sebastian Bonhoeffer at ETH Zurich and Prof. Narendra Dixit’s group at the Indian Institute of Science in Bangalore, we modelled within-host evolution of drug resistance and immune escape in viral pathogens. A common theme was the initial stochastic appearance and spread of rare adaptive mutants. This work combined techniques of ordinary differential equations, stochastic process analysis, and computational simulations.
Although currently dormant in our group, work on viral infection dynamics remains of interest!
- Alexander & Bonhoeffer, Epidemics, 2012
- Nagaraja, Alexander et al., Epidemics, 2016
Microbial life history
One long-standing interest is how microbial life history influences evolution. In past work, we have specifically examined the impact of mutations and drug inhibition affecting different steps of viral replication cycles.
- Alexander & Wahl, Evolution, 2008
- Alexander & Bonhoeffer, Epidemics, 2012
Other past projects
Past areas of research that are no longer active in our group include:
- modelling the role of regulatory T cells in autoimmune disease (with Prof. Lindi Wahl)
- developing methods for phylodynamic inference with age-dependent mortality (with Prof. Amaury Lambert and Prof. Tanja Stadler)
This article was published on

