Research overview
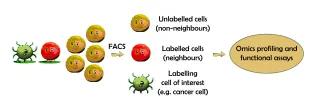
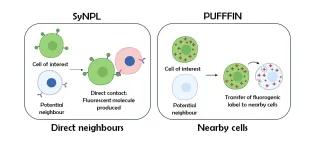
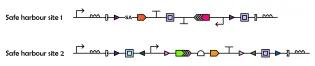
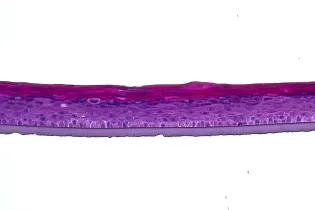
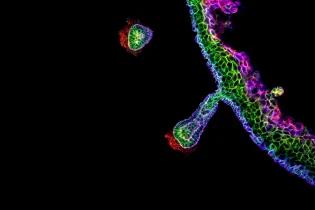
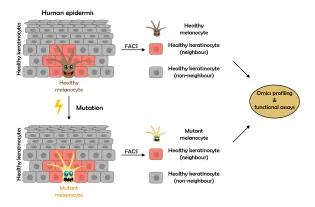
Cell-cell interactions profoundly influence cellular decision-making during development, in homeostasis, and during the onset and progression of disease. However, identifying these interactions and decoding their downstream effects remains technically challenging due to a shortage of relevant analytical approaches.