View the list of our publications.
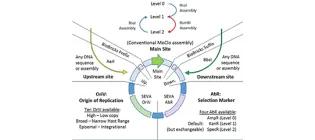
Joint Universal Modular Plasmids (JUMP): A flexible and comprehensive platform for synthetic biology
Valenzuela-Ortega, M. & French, C.E. (2019). https://doi.org/10.1101/799585
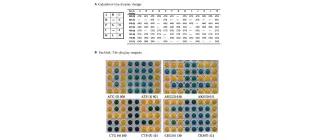
ParAlleL: a novel population-based approach to biological logic gates
Millacura, F. A., Largey, B., & French, C. E. (2019) Frontiers in Bioengineering and Biotechnology, 7.
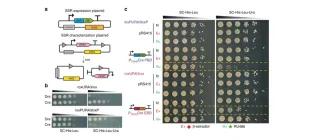
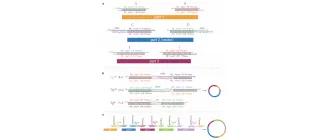
Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods
Liu, W., Liu, W., Luo, Z., Wang, Y., Pham, N.T., Tuck, L., Pérez-Pi, I., Liu, L., Shen, Y., French, C., Auer, M. and Marles-Wright, J. (2018). Nature communications, 9(1), 1936.
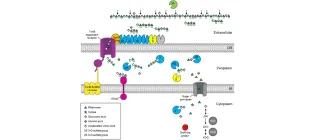
The enzymatic ulvan depolymerisation system from the alga-associated marine flavobacterium Formosa agariphila
Salinas, A., & French, C. E. (2017). Algal Research, 27, 335-344.
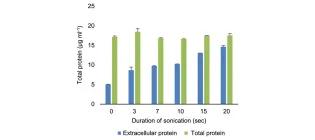
Data for discriminating dead/live bacteria in homogenous cell suspensions and the effect of insoluble substrates on turbidimetric measurements
Duedu, K. O., & French, C. E. Data in Brief, 12, 169-174.
Characterization of a Cellulomonas fimi exoglucanase/xylanase-endoglucanase gene fusion which improves microbial degradation of cellulosic biomass
Duedu, K. O., & French, C. E. (2016). Enzyme and Microbial Technology, 93, 113-121.
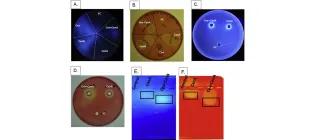
Citrobacter freundii as a test platform for recombinant cellulose degradation systems
Lakhundi, S. S., Duedu, K. O., Cain, N., Nagy, R., Krakowiak, J., & French, C. E. (2016). Letters in applied microbiology, 64(1), 35-42.
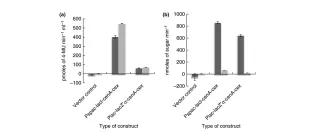
BBF RFC 104: BrickClip–rapid assembly of multiple RFC10 BioBricks
Trubitsyna, M., Chan, K., Cai, Y., Elfick, A., & French, C. (2015).
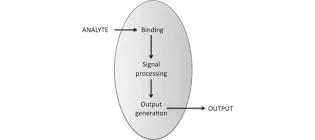
Beyond genetic engineering: Technical capabilities in the application fields of biocatalysis and biosensors
French, C. E., Horsfall, L., Barnard, D. K., Duedu, K. O., Fletcher, E., Joshi, N., … & Radford, D. (2015). In Synthetic Biology (pp. 113-137). Springer International Publishing.
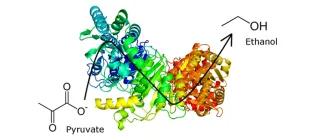
Fusion of pyruvate decarboxylase and alcohol dehydrogenase increases ethanol production in Escherichia coli
Lewicka, A. J., Lyczakowski, J. J., Blackhurst, G., Pashkuleva, C., …, Horsfall, L., Elfick, A., & French, C. E. (2014). ACS Synth. Biol., 3 (12), 976–978.
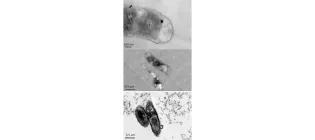
Enhanced resistance to nanoparticle toxicity is conferred by overproduction of extracellular polymeric substances
Joshi, N., Ngwenya, B. T., & French, C. E. (2012). Journal of hazardous materials, 241, 363-370.

Synthetic biology and the art of biosensor design
French, C. E., de Mora, K., Joshi, N., Elfick, A., Haseloff, J., & Ajioka, J. (2011, December). The science and applications of synthetic and systems biology: Workshop summary (pp. 178-201). Washington DC: National Academies Press.
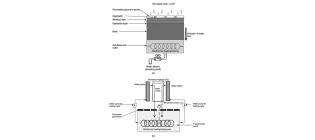
The synergy of permeable pavements and geothermal heat pumps for stormwater treatment and reuse
Tota‐Maharaj, K., Scholz, M., Ahmed, T., French, C. E., & Pagaling, E. (2010). Environmental technology, 31(14), 1517-1531.
Synthetic biology and biomass conversion: a match made in heaven?
French, C. E. (2009). Journal of the Royal Society Interface, rsif20080527.

Diversity and distribution of hemerythrin-like proteins in prokaryotes
French, C. E., Bell, J. M., & Ward, F. B. (2008). FEMS microbiology letters, 279(2), 131-145.
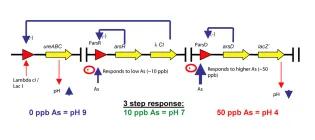
Arsenic biosensor: a step further
French, C.E., Nicholson, J., Bizzari, F., Aleksic, J., Cai, Y., Seshasayee, S. L., ... & Ma, H. (2007). BMC Systems Biology, 1(1), S11.
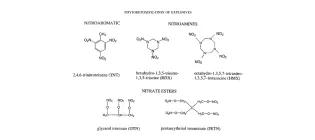
Engineering plants for the phytodetoxification of explosives
Rosser, S. J., French, C.E., & Bruce, N. C. (2001). In Vitro Cellular & Developmental Biology-Plant, 37(3), 330-333.
Microbial transformations of explosives
Rosser, S. J., Basran, A., Travis, E. R., French, C.E., & Bruce, N. C. (2001). Advances in applied microbiology, 49, 1-35.
Cloning of the sth gene from Azotobacter vinelandii and construction of chimeric soluble pyridine nucleotide transhydrogenases
Boonstra, B., Björklund, L., French, C.E., Wainwright, I., & Bruce, N. C. (2000). FEMS microbiology letters, 191(1), 87-93.

Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase
French, C. E., Rosser, S. J., Davies, G. J., Nicklin, S., & Bruce, N. C. (1999). Nature biotechnology, 17(5), 491-494.
The Degradation of Nitrate Ester Explosives and TNT by Enterobacter cloacae PB2
French, C. E., Binks, P. R., Rathbone, D. A., Rosser, S. J., Williams, R. E., Nicklin, S., & Bruce, N. C. (1999). Novel Approaches for Bioremediation of Organic Pollution (pp. 59-69). Springer US.
Aerobic degradation of 2, 4, 6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase
French, C. E., Nicklin, S., & Bruce, N. C. (1998). Applied and Environmental Microbiology, 64(8), 2864-2868.
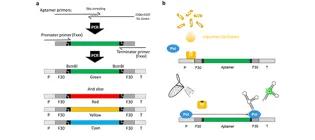
TOBIAS: Transcription-Only Biological Information Algorithmic Systems as a novel cell-free approach for Synthetic Biology.
Millacura, F.A., Li, M., Valenzuela-Ortega, M., & French C.E. (2019).
Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals.
Wan, X., Volpetti, F., Petrova, E., French, C., Maerkl, S. J., & Wang, B. (2019). Nature chemical biology, 1.
Characterisation of novel biomass degradation enzymes from the genome of Cellulomonas fimi.
Kane, S. D., & French, C. E. (2018). Enzyme and microbial technology, 113, 9-17.
Two-colour fluorescence fluorimetric analysis for direct quantification of bacteria and its application in monitoring bacterial growth in cellulose degradation systems.
Duedu, K. O., & French, C. E. (2017). Journal of Microbiological Methods, 135, 85-92.

Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome.
Shen, Y., Wang, Y., Chen, T., Gao, F., Gong, J., Abramczyk, D., & 33 others including French, C. E., … & Luo, Y. (2017). Science, 355(6329), eaaf4791
Use of mariner transposases for one-step delivery and integration of DNA in prokaryotes and eukaryotes by transfection
Trubitsyna, M., Michlewski, G., Finnegan, D. J., Elfick, A., Rosser, S. J., Richardson, J. M., & French, C. E. (2017). Nucleic Acids Research, 1.
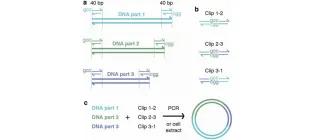
PaperClip: A Simple Method for Flexible Multi-Part DNA Assembly.
Trubitsyna, M., Liu, C. K., Salinas, A., Elfick, A., & French, C. E. (2016). Synthetic DNA: Methods and Protocols, 111-128.
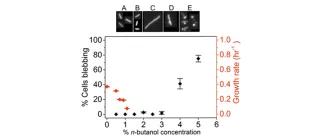
Characterization of the effects of n-butanol on the cell envelope of E. coli
Fletcher, E., Pilizota, T., Davies, P. R., McVey, A., & French, C. E. (2016). Applied microbiology and biotechnology, 100(22), 9653-9659.
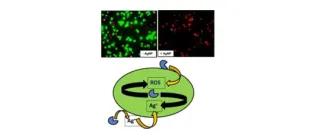
Use of bioreporters and deletion mutants reveals ionic silver and ROS to be equally important in silver nanotoxicity.
Joshi, N., Ngwenya, B. T., Butler, I. B., & French, C. E. (2015). Journal of hazardous materials, 287, 51-58.
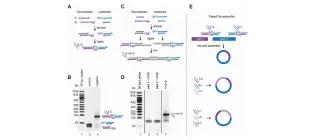
PaperClip: rapid multi-part DNA assembly from existing libraries.
Trubitsyna, M., Michlewski, G., Cai, Y., Elfick, A., & French, C. E. (2014). Nucleic acids research, gku829.
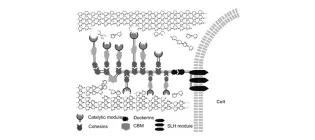
Synthetic biology for biomass conversion
French, C. E., Barnard, D. K., Fletcher, E., Kane, S. D., Lakhundi, S. S., Liu, C. K., & Elfick, A. (2013). New and future developments in catalysis. Elsevier, Amsterdam, 115-140., 115-140
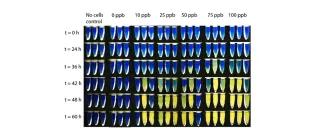
A pH-based biosensor for detection of arsenic in drinking water
Mena-Benitez, G. L., Gandia-Herrero, F., Graham, S., Larson, T. R., McQueen-Mason, De Mora, K., Joshi, N., Balint, B. L., Ward, F. B., Elfick, A., & French, C. E. (2011). Analytical and bioanalytical chemistry, 400(4), 1031-1039.

Polymer-induced phase separation in suspensions of bacteria
Schwarz-Linek, J., Dorken, G., Winkler, A., Wilson, L. G., Pham, N. T., French, C. E., … & Poon, W. C. K. (2010). EPL (Europhysics Letters), 89(6), 68003.

Novel approaches to biosensors for detection of arsenic in drinking water.
Joshi, N., Wang, X., Montgomery, L., Elfick, A., & French, C. E. (2009). Desalination, 248(1-3), 517-523.
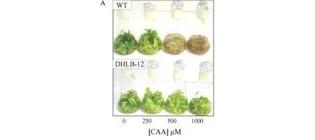
Engineering a catabolic pathway in plants for the degradation of 1, 2-dichloroethane.
Mena-Benitez, G. L., Gandia-Herrero, F., Graham, S., Larson, T. R., McQueen-Mason, S. J., French, C. E., … & Bruce, N. C. (2008). Plant physiology, 147(3), 1192-1198.
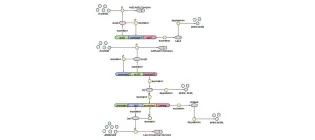
Development of a novel biosensor for the detection of arsenic in drinking water
Aleksic, J., Bizzari, F., Cai, Y., Davidson, B., De Mora, K., Ivakhno, S., … & French, C. E. (2007). IET Synthetic biology, 1(1), 87-90.
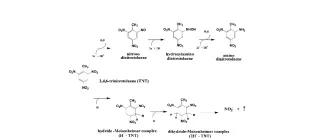
Uptake and metabolism of TNT and GTN by plants expressing bacterial pentaerythritol tetranitrate reductase
Hannink, N. K., Rosser, S. J., French, C. E., & Bruce, N. C. (2003). Water, Air, & Soil Pollution: Focus, 3(3), 251-258.

Phytodetoxification of TNT by transgenic plants expressing a bacterial nitroreductase
Hannink, N., Rosser, S. J., French, C. E., Basran, A., Murray, J. A., Nicklin, S., & Bruce, N. C. (2001). Nature biotechnology, 19(12), 1168-1172.
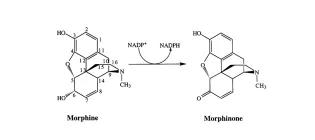
Mechanistic studies of morphine dehydrogenase and stabilization against covalent inactivation
Walker, E.H., French, C.E., Rathbone, D.A., & Bruce, N.C. (2000). Biochemical Journal, 345(3), 687-692.
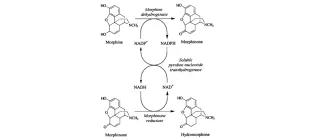
Cofactor regeneration by a soluble pyridine nucleotide transhydrogenase for biological production of hydromorphone
Boonstra, B., Rathbone, D. A., French, C.E., Walker, E. H., & Bruce, N. C. (2000). Applied and environmental microbiology, 66(12), 5161-5166.
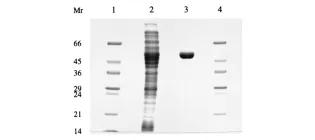
The udhA gene of Escherichia coli encodes a soluble pyridine nucleotide transhydrogenase
Boonstra, B., French, C. E., Wainwright, I., & Bruce, N. C. (1999). Journal of bacteriology, 181(3), 1030-1034.

Crystallization and preliminary diffraction studies of pentaerythritol tetranitrate reductase from Enterobacter cloacae PB2
Moody, P. C., Shikotra, N., French, C.E., Bruce, N. C., & Scrutton, N. S. (1998). Acta Crystallographica Section D: Biological Crystallography, 54(4), 675-677.
This article was published on

