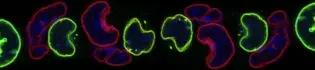
In eukaryotes, DNA is packaged into a nucleoprotein complex known as chromatin. There are two main different types of chromatin, euchromatin and heterochromatin. Euchromatin is characteristically more accessible and consists of actively transcribed genomic regions that replicate early during S-phase. Conversely, heterochromatin is tightly packed and includes either repressed genes or genomic areas essential for organising the nuclear chromatin architecture. Heterochromatin replicates during late S-phase. Euchromatin and heterochromatin are distinguished by the combination of specific post-translational modifications (PTMs) on the histones. This epigenetic profile, the timing of replication, subnuclear positioning and inter-chromatin contacts (chromatin three-dimensional organisation) are interconnected features that converge into the determination of the gene expression profile and, consequently, cellular identity.
DNA replication timing, epigenetic and chromatin architecture: how to shape cell identity
During DNA replication, the histone PTMs are diluted following the doubling of the DNA and the incorporation of newly synthesised, naïve histones. Reinstating the histone PTMs characteristic of the specific genomic region is essential to preserve the identity and genomic stability in the daughter cells. Errors in this process can indeed lead to genomic instability, cellular dysfunction, and contribute significantly to diseases such as cancer or age-related tissue and organ degeneration. The mechanisms that ensure the precise reestablishment of epigenetic identity, particularly for heterochromatin, are poorly understood.
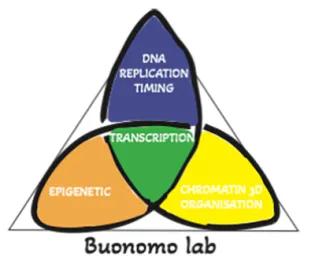
Our group has identified a key regulator of the replication-timing program, RIF1. The next big question is what the biological function of this highly conserved program is. A long-standing hypothesis proposes that replication at different stages of S-phase aids the maintenance of the epigenetic identity of various genomic regions. We are investigating this hypothesis and, more widely, the molecular relationships between the different types of chromatin (epigenetic identity), the timing of their replication and their three-dimensional organisation in the nucleus.
We are particularly interested in different forms of heterochromatin, including the inactive X chromosome, heterochromatin associated with the nuclear lamina (LADs), and endogenous retroviruses. Our goal is to discover and distinguish universal mechanisms from those that are specific to certain types of heterochromatin.
We are also investigating the hypothesis that the replication-timing program could contribute to coordinating transcription and replication. Conflicts between transcription and replication are potential sources of genomic instability and mutations that weaken this control contribute to pathology such as neurodegeneration or cancer.
Related links
This article was published on